Catalog summary
RELEASED: 2022-07-09
GENOME ASSEMBLY GRCh38.p13
PUBLICATIONS 5,848
ASSOCIATIONS 398,342
Search For:
Blog Categories
Trait annotation in the GWAS Catalog 2021-08-03T00:00:00.000Z By Elliot Sollis
Each study in the GWAS Catalog investigates the association between variants in the human genome and a particular trait or phenotypic characteristic.
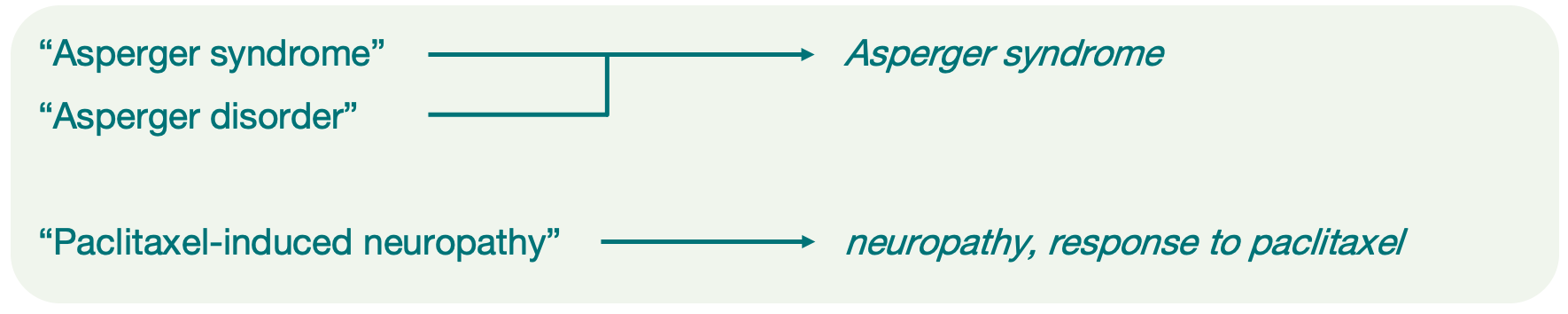
For each study, we annotate the trait in two ways:
-
A reported trait that reflects the author’s description of the disease or phenotypic characteristic under investigation. This is a free text description and sometimes different studies might use a different wording to describe similar traits or to capture more nuanced distinctions. Reported traits can also include multiple component traits, depending on the study design.
-
One or more trait terms from the Experimental Factor Ontology (EFO) that represent phenotypic characteristics in a more standardised way. These terms make studies on similar traits easier for users to find and compare. For multifaceted traits, each component is represented by a separate term.
Annotating different study types
A. Single-trait studies
The vast majority (>80%) of studies in the GWAS Catalog only analyse a single trait. We annotate these studies with a single EFO term.
Some common examples include:
- Standard case-control studies comparing individuals with a disease or phenotypic characteristic, versus controls individuals without that trait.

- Quantitative studies looking at a single measurement

In these examples, any reported variants are clearly and straightforwardly associated with that single trait.
B. Multi-trait studies
In about 10% of studies, there are multiple traits of interest that are analysed simultaneously. We annotate these studies with multiple EFO terms separated by a comma, indicating that any significant variants reported in the study are associated with both traits in some way.
Some common examples include:
- Studies comparing individuals with two comorbid diseases (or other traits), versus control individuals who have neither disease. In the reported trait we write “Disease 1 and Disease 2”. In the trait we list multiple EFO terms separated by a comma.

- Studies comparing individuals with either of two diseases (or other traits), versus control individuals who have neither disease. Often these are two traits that are hypothesised to have some common underlying genetic factors (pleiotropy). In the reported trait we write “Disease 1 or Disease 2”. In the trait we list multiple EFO terms separated by a comma.

In these examples, any significant variants reported are associated with either or both of the annotated traits. If a user is searching for associations with either trait term, then these results will come up in their search.
C. Studies with a background trait
Finally, about 6% of studies analyse only one main trait of interest, but in the context of a background trait that is shared by all of the participants in the study.
Some examples include:
- Studies comparing cases and controls of one disease, but only within a sample of people who have another disease. In the reported trait we write “Disease 1 in Disease 2”. In the past, we listed multiple EFO terms in the trait column, as for scenario B.

- Quantitative studies analysing a measurement in the context of a disease. In the reported trait we write “Measurement in Disease”. In the past, we listed multiple EFO terms in the trait column, as for scenario B.
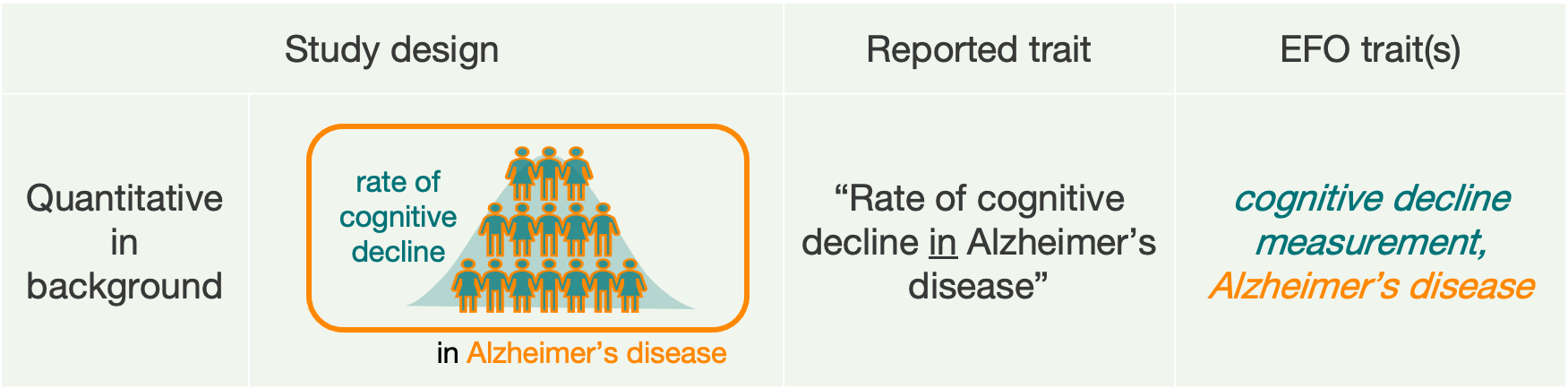
In these examples, any reported variants are associated with the main trait, but not with the background trait.
Room for improvement
In the past, we annotated background trait studies (scenario C) in the same way as we have for multi-trait studies (scenario B): with multiple EFO terms listed together in the trait field, separated by commas.
This had some benefits:
-
We were able to indicate that the study has something to do with the background trait - e.g. a particular association with allergic rhinitis may only hold true in asthmatics.
-
Users could search for associations with a trait, as well as associations that are found in the context of that trait as a background characteristic. Both kinds of associations might be relevant to users working in a particular field.
However, there were some are also some disadvantages:
-
It was not possible to tell that a particular EFO term was a background trait, without looking at the reported trait field. This complicated analysis particularly when accessing the data programmatically.
-
When searching for a trait term (e.g. asthma), there was no easy way to distinguish which studies were direct associations with asthma, and which have asthma as the background trait, without reading the reported trait for each one.
-
The overall study and association numbers provided for some traits could be misleading since they included studies and associations where the trait of interest was a background trait.
Changes to trait annotation
To make our trait annotations more informative, we have added an additional background trait field to the GWAS Catalog database.
We have moved all EFO terms related to background traits to this new field, and removed them from the original trait field:

Changes to the website
We have also updated our web interface in order to display the restructured background trait information.
Data tables
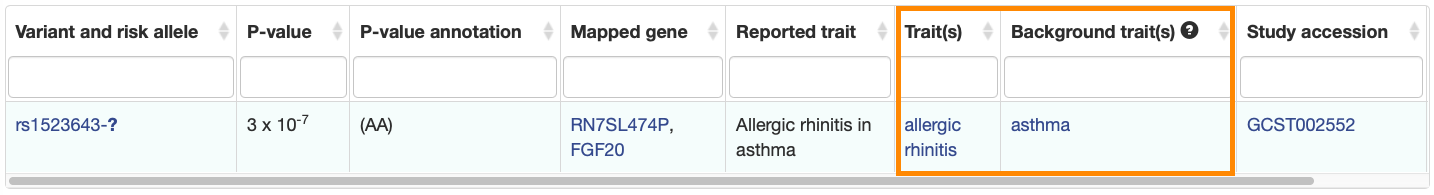
New columns have been added to the Associations and Studies tables to clearly indicate the main and background traits.
Associations:
Studies:
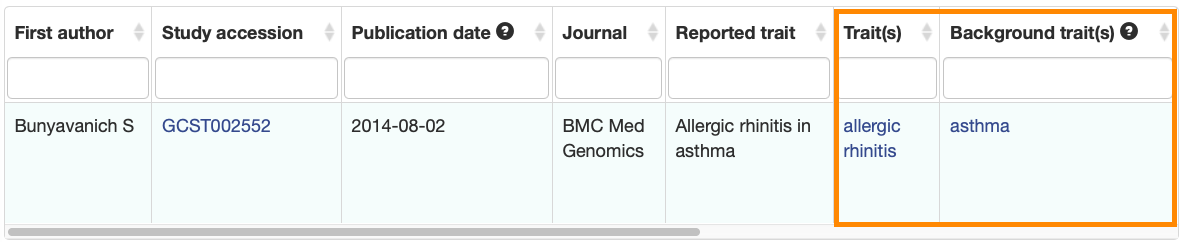
The improved tables will be displayed on all Publication, Study, Trait, Variant, Gene and Region pages.
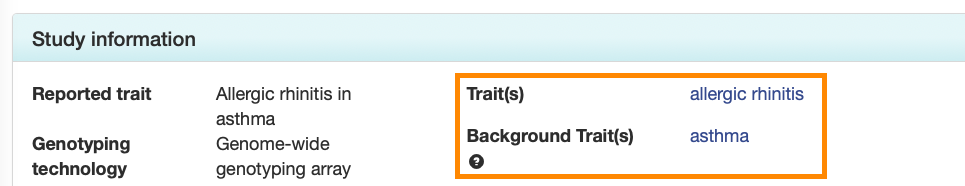
Study pages
Each Study page also displays the main and background traits in separate fields in the Study Information panel.
Trait pages
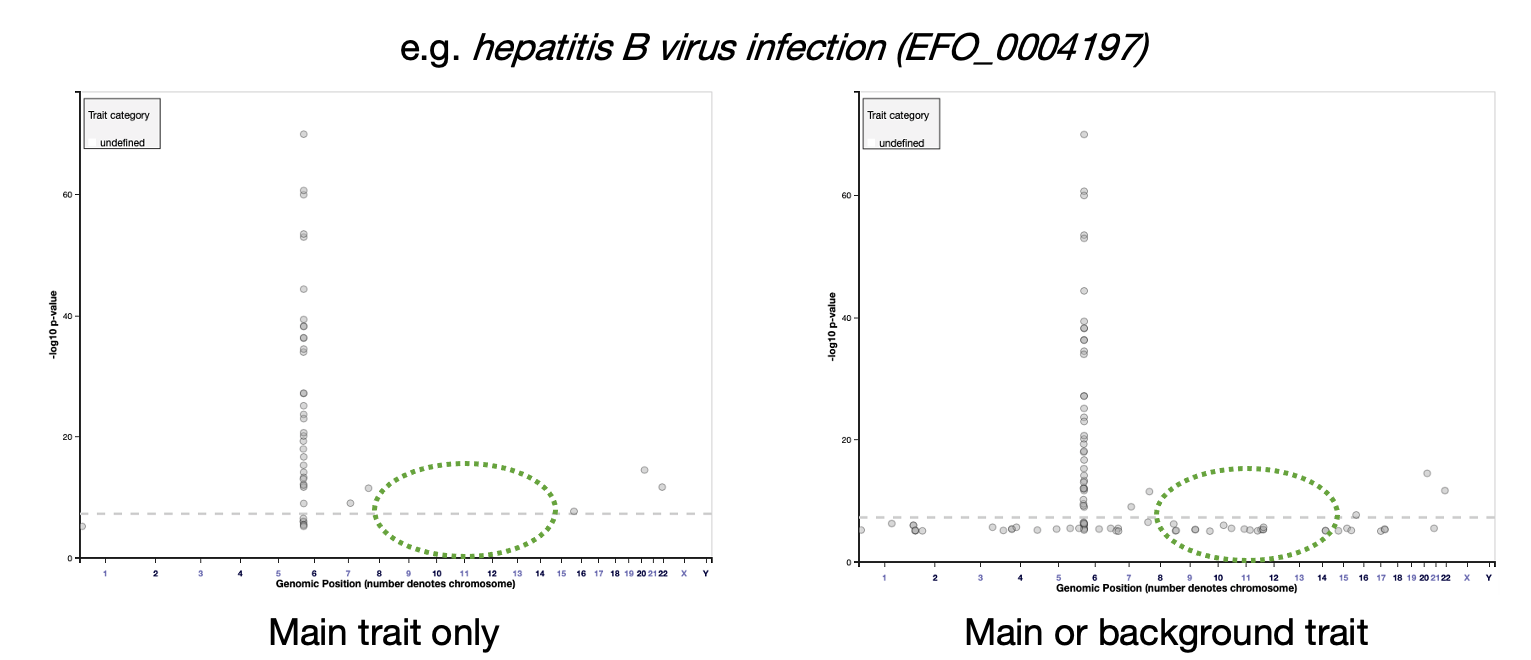
The Trait page has been updated so that background trait studies and associations are no longer shown by default. For example, the asthma page only shows associations with asthma itself, and not associations with others traits in asthma.
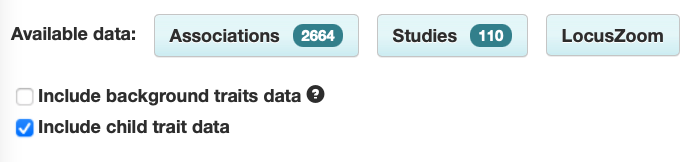
Shows associations with:
- asthma
[other traits] in asthma
However, users can select the box to include background trait data if they wish:
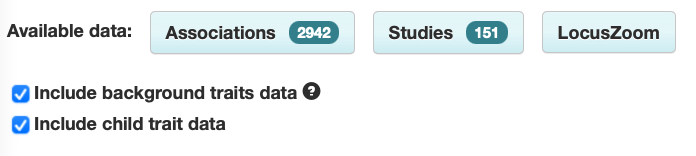
Shows associations with:
- asthma
- [other traits] in asthma
Some trait terms may only be used in the GWAS Catalog to annotate background traits. These terms will continue to have their own Trait page, but no associations or studies will be displayed under the default view. For example, autoimmune pancreatitis type 1 (EFO_1000780) currently appears in the Catalog only as a background trait for one study (“Lachrymal/Salivary gland lesion in type 1 autoimmune pancreatitis”), so no associations with autoimmune pancreatitis type 1 are displayed by default:
The association plot on each Trait page will now display only main trait associations by default, but background trait data can be added by selecting the box:
Downloads
In the full Catalog downloads, the MAPPED_TRAIT and MAPPED_TRAIT_URI columns will now only show EFO terms for main traits. Background trait terms have been removed from these columns.
In the newest version of the download (v1.0.3), new columns have been added for the background trait: MAPPED BACKGROUND TRAIT and MAPPED BACKGROUND TRAIT URI. This is not included in earlier versions of the download.
API
For users of the GWAS Catalog API, searching for “associationByEfoTrait” will now return only main trait associations.
There is currently no background trait field available in the API, but we plan to add this feature in the future.
Expected scope of the changes
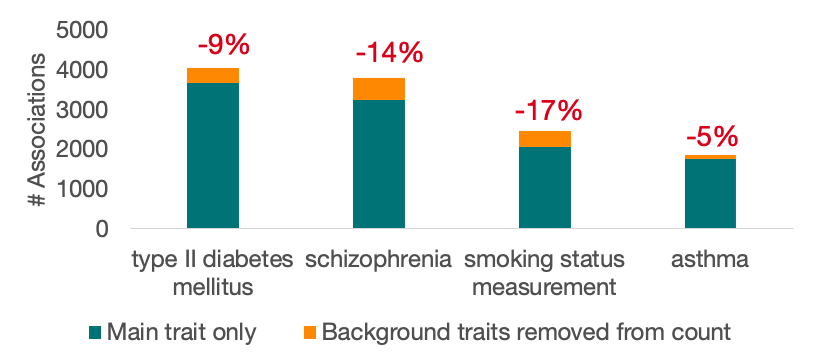
We reviewed all of the studies in the Catalog and identified just over 1000 studies (about 6% of the total) with background traits that have now been moved to the new field. These studies contain around 10,000 associations (about 4% of all associations in the Catalog) which have also been reannotated.
The changes affect some trait terms more than others. Here are some of the terms that will see the greatest change in the number of annotated Associations:
Questions and feedback
If you have any questions or comments about this change, please contact us as gwas-info@ebi.ac.uk.