Catalog summary
RELEASED: 2022-07-09
GENOME ASSEMBLY GRCh38.p13
PUBLICATIONS 5,848
ASSOCIATIONS 398,342
Search For:
Blog Categories
Our new resource update paper is out in Nucleic Acids Research! 2024-11-20T00:00:00.000Z By GWAS Catalog team
We're happy to announce the GWAS Catalog’s latest resource update paper in Nucleic Acids Research!

Here’s a hint of what you can find inside:
The GWAS-SSF standard for full genome-wide summary statistics data and metadata has been fully implemented in our submission pipeline. More than 30k files have been submitted containing ALL mandatory data & metadata fields, on top of 50k files in the previous format. We’ve also scaled our harmonisation pipeline to keep up with the increased rate of data submission - now 92% of all summary statistics with the necessary data content have a harmonised version available.
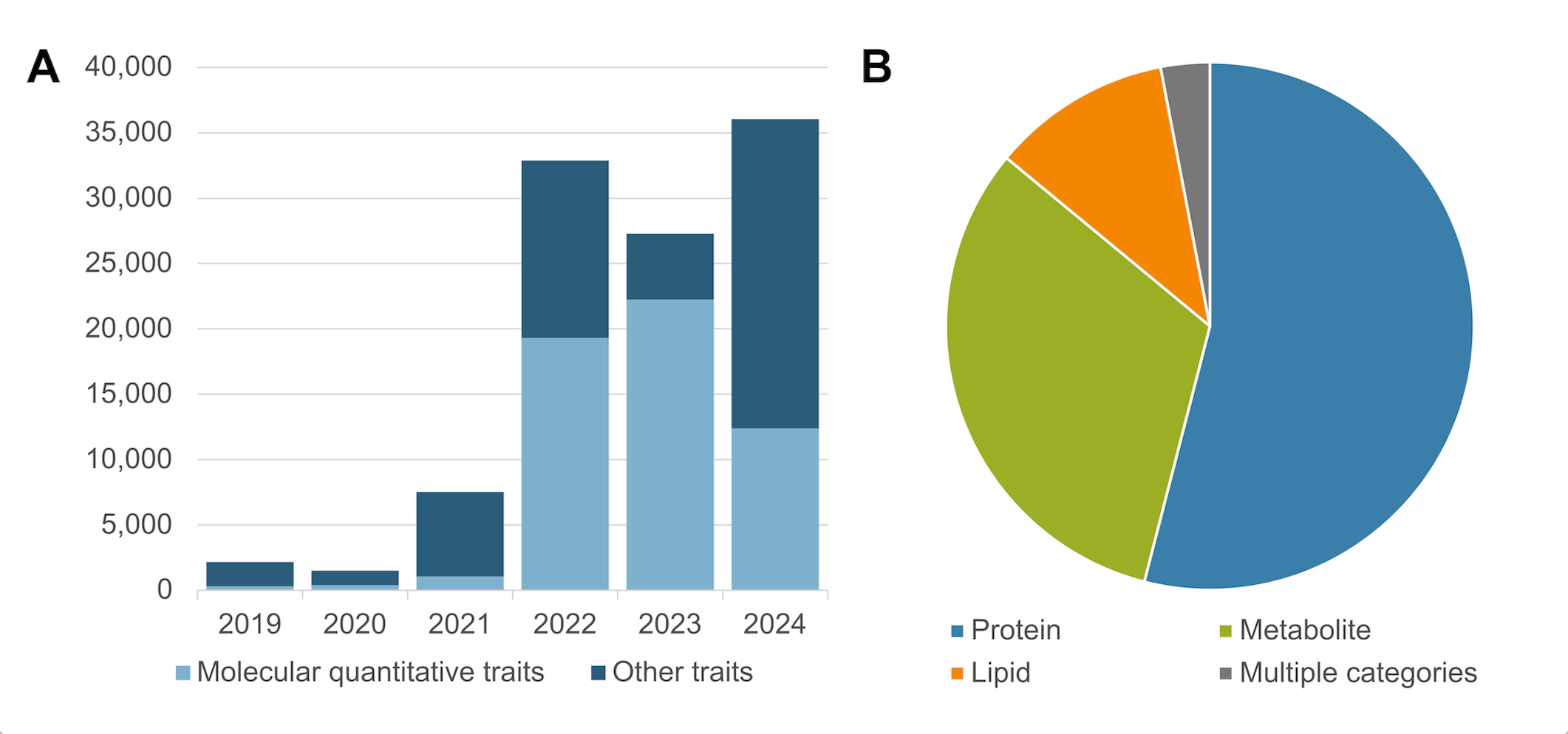
We have seen a huge growth in molecular quantitative trait GWAS (proteomic, lipidomic & metabolomic measurements) and are working with our colleagues at the Experimental Factor Ontology to improve mapping to chemical entities.
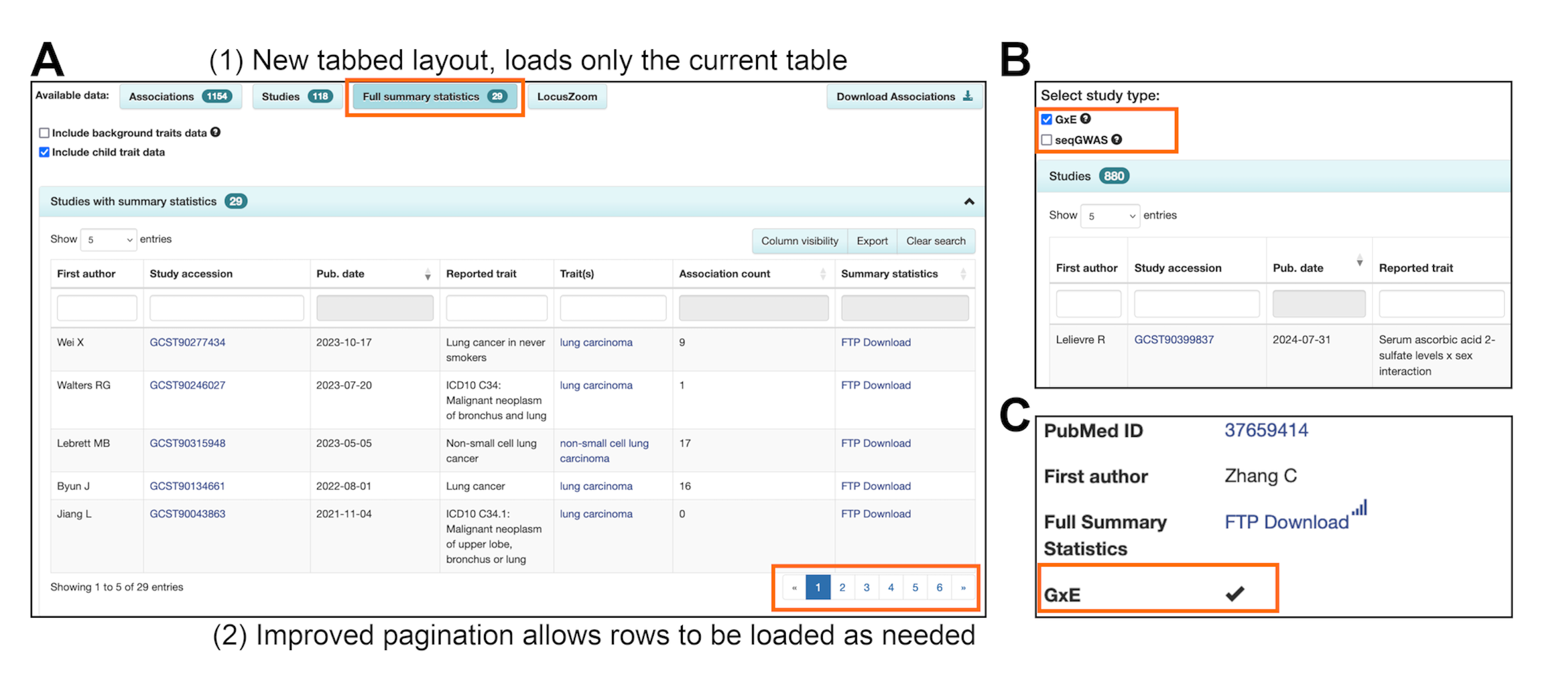
New features enable faster page loading and allow you to easily find GxE studies in our website and download files, preparing the way for anticipated growth in GxE data is anticipated to grow as the scale of exposome data expands. Thanks to funding from the Human Ecosystems Transversal Theme at EMBL-EBI to enable this new feature!
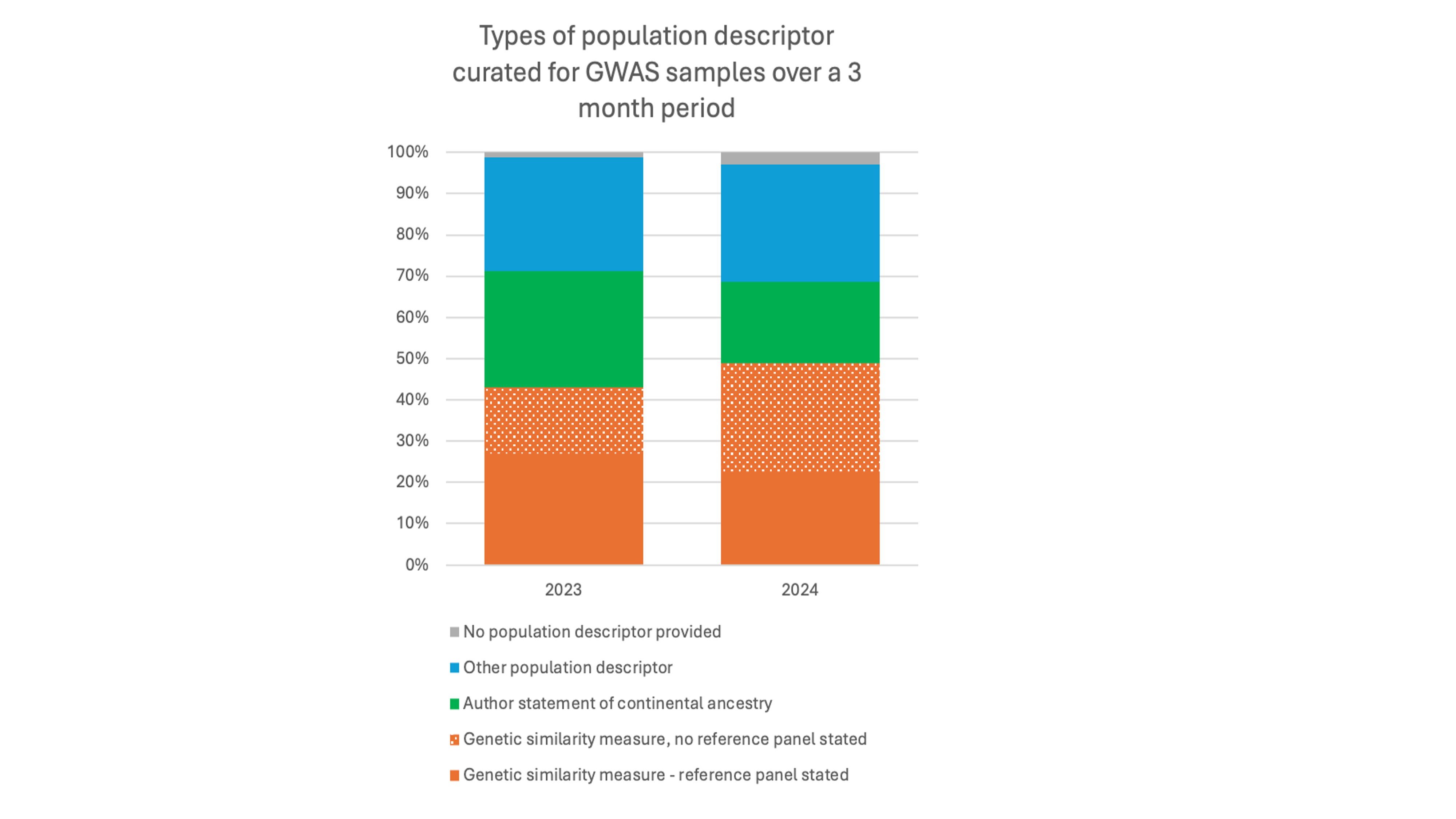
The lack of diversity in genomics data continues to be an issue. We share our thoughts on how to enable monitoring of diversity and inclusion in GWAS studies while taking care that the language we use to describe populations does not lead to misuse. New documentation and terminology (labels instead of categories) guide our users towards correct interpretation.
Our analysis found that only around 50% of GWAS samples reported in the literature are being defined by genetic similarity measures, of those less than half clearly described a reference panel.
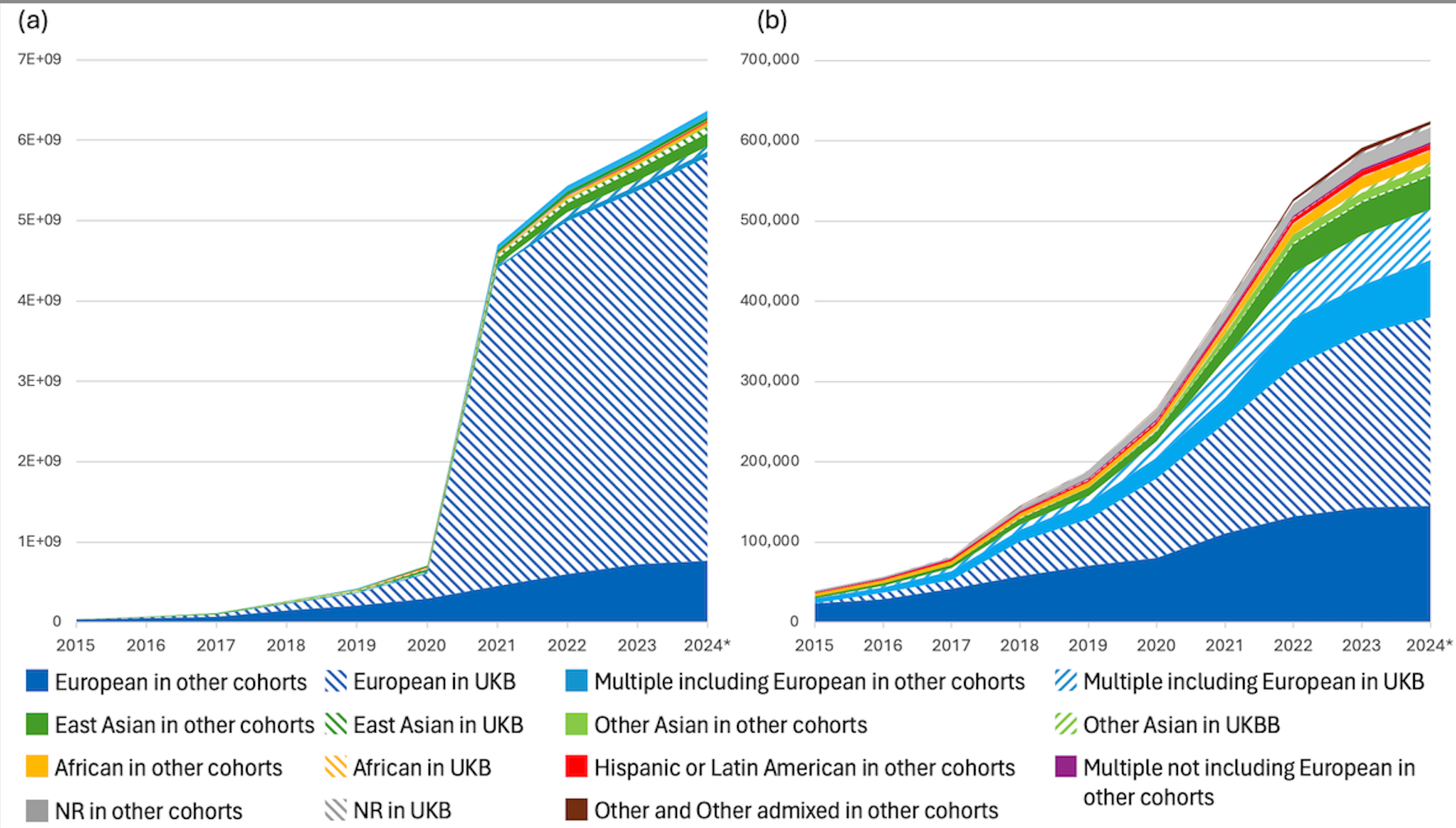
UKB is still the most used cohort in GWAS, accounting for around 30% of all studies in the Catalog! Unfortunately 70% of these restrict analysis to samples with the European population label, losing a significant amount of diversity. New inclusion of cohort names in Catalog data now enables this analysis, and in future linking out to rich cohort metadata.
Finally, we are now recognised as a Global Biodata Core Resource and Elixir Core Data resource!

Read the full article, Cerezo et al 2024, open access at NAR.
Comments, feedback…? Contact us via gwas-info@ebi.ac.uk