Catalog summary
RELEASED: 2022-07-09
GENOME ASSEMBLY GRCh38.p13
PUBLICATIONS 5,848
ASSOCIATIONS 398,342
Search For:
Blog Categories
Streamlining GWAS Summary Statistics Data Interpretation with Improved YAML Metadata Feburary 22, 2024 By Yue Ji and Laura Harris
Having clear and accessible metadata is essential for enhancing data interpretation and ensuring its reusability. In the case of Genome-Wide Association Studies (GWAS), having a standardized and easy-to-understand format for documenting study metadata is crucial. In the GWAS Catalog, metadata associated with full genome-wide summary statistics files is accessible via multiple routes - searchable in the main Catalog via the website and REST API, and additionally via a text file in YAML format, contained in the same directory as the data file.
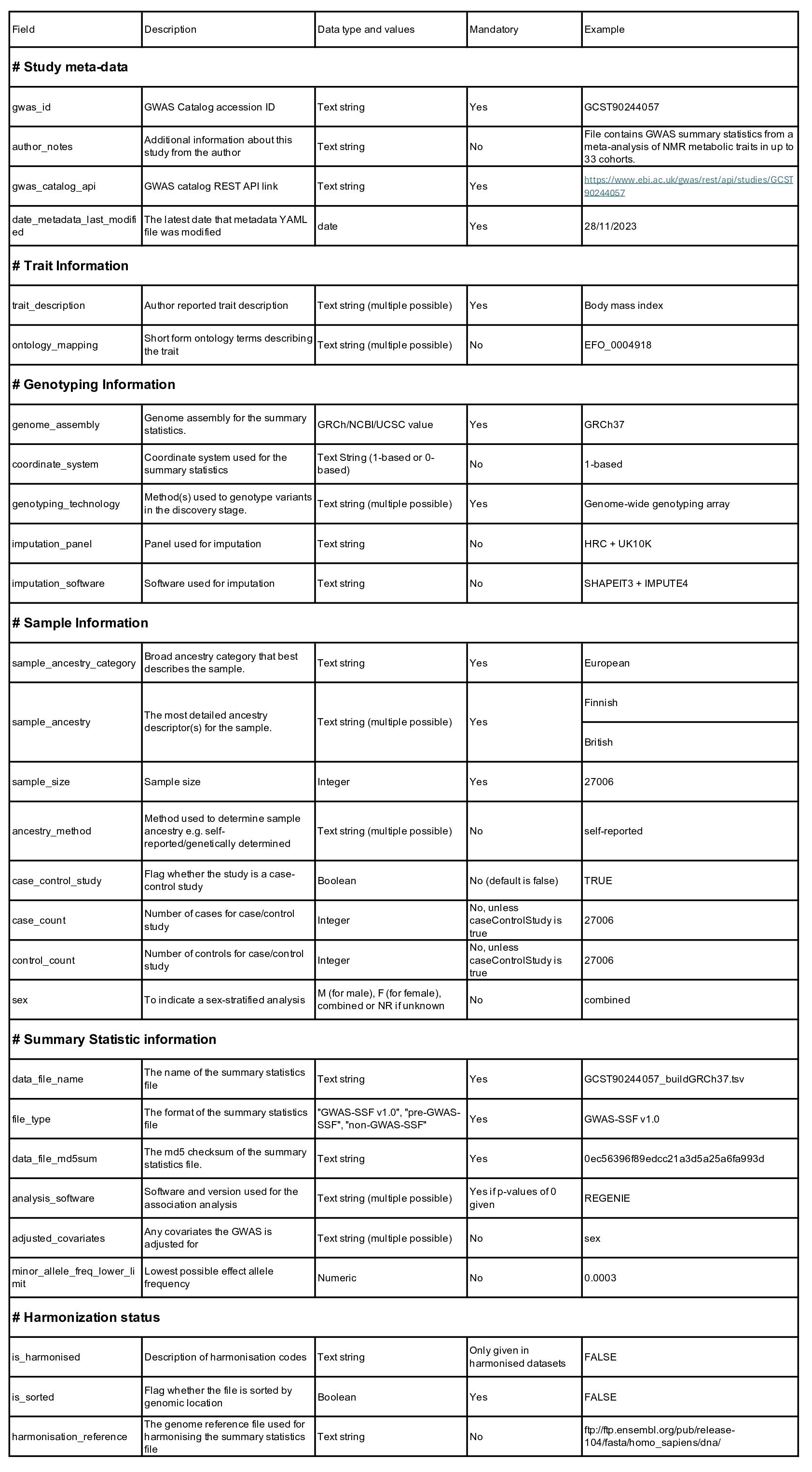
We have identified key fields that should be included in the metadata for a comprehensive understanding, interpretation, and subsequent reuse of GWAS studies. Mandatory fields document essential study details and optional fields cater to specific study designs (such as case/control counts in case-control studies), and provide supplementary analysis information. You can find detailed information about these fields in Table 1 and in our recent preprint (Hayhurst et al 2023 https://www.biorxiv.org/content/10.1101/2022.07.15.500230v4).
In our latest release, we've introduced several updates to the GWAS metadata YAML file format. The YAML format, known it's user-friendliness, provides a combination of structured information and ease of use for humans and computers. The latest changes aim to enhance the human readability of the files with a clearer, more unambiguous, and user-friendly experience without compromising the file's compatibility with automated systems. Here’s an overview of the significant enhancements:
- Reordering and Grouping of Fields:
- The fields within the YAML files have been reorganized and grouped by related topics, each marked with a group name starting with '#' (group names are listed in Table 1). This organization helps users to quickly locate specific fields by group name, such as '# Sample Information', making the file more navigable for humans. Importantly, this structural refinement does not affect the way machines parse and interpret the data.
- Clear Indentation for Hierarchical Relationships:
- Indentation has been introduced to visually represent the relationships between different data fields. This visual aid improves the readability of the files, making it easier for users to discern the levels of data organization and relationships between various fields. In the meantime, this indentation does not impact the machine processing of the file.
- Renaming 'data_last_modified' to 'date_metadata_last_modified':
- To provide clearer context, the 'data_last_modified' field has been renamed to 'date_metadata_last_modified'. This change emphasizes that the timestamp refers to the last modification date of the metadata, not the summary statistics data. It aids users in tracking any changes or updates in the metadata over time.
- Addition of 'Ancestry Category' Field:
- We have introduced a new 'ancestry category' field in addition to the existing 'ancestry' field. This new feature provides a broad overview of the ancestry category that best describes the sample, allowing for a more generalized view of the sample's ancestry. This new field also ensures consistency and facilitates comparative analysis across different studies, in line with the rest of the Catalog.
These updates reflect our commitment to improving the user experience while ensuring the YAML files maintain their structural integrity for computer readability. Our focus continues to be on making GWAS metadata more accessible, interpretable, and useful for researchers.
Table 1. Metadata field definitions
Questions and feedback
Questions or comments about this change? Please contact us as gwas-info@ebi.ac.uk.