Catalog summary
RELEASED: 2022-07-09
GENOME ASSEMBLY GRCh38.p13
PUBLICATIONS 5,848
ASSOCIATIONS 398,342
Search For:
Blog Categories
Ten years of the GWAS Catalog – Past, present and future 2018-06-28T00:00:00.000Z By Laura Harris (GWAS Catalog) and Astrid Gall (Ensembl Outreach)
This year marks a special anniversary for the GWAS Catalog, as we have reached ten years since our launch in 2008. The GWAS Catalog is a widely used publicly available resource of all published human genome wide association studies (GWAS) and association results. Each GWAS study contains a wealth of information which is effectively inaccessible to researchers and clinicians without them spending a lot of time undertaking regular systematic reviews of the literature. This is where we can help you!
The project is a collaboration between the National Human Genome Research Institute (NHGRI) in Bethesda, Maryland, USA, and the European Molecular Biology Laboratory – European Bioinformatics Institute (EMBL-EBI) in Hinxton, UK, where the majority of our curators and all of our software developers are based

At EMBL-EBI, we share the building with the Ensembl team and work very closely with them – which is very handy for both teams!
Our curators extract genotype-phenotype associations from all published GWAS studies, along with study information to allow you to interpret the data accurately. We make the data structured, searchable and visualisable, annotate it and integrate it with other resources including Ensembl. Each association with a standard variant identifier (rsID) enters our automated Ensembl mapping pipeline which provides the genomic annotation, i.e. the location on the chromosome and cytogenetic band as well as the mapped genes. We use this pipeline to check for errors as part of our quality control and display the information in our interface.
Our colleagues at Ensembl benefit from the GWAS Catalog as they import variant and phenotype data from it. More than 74,000 human variants associated with phenotype data from the GWAS catalog were included in the latest Ensembl release (e92); and the number is increasing continuously!
[video]
Over the last ten years, the GWAS Catalog has grown, from a single published GWAS on age-related macular degeneration, into the primary source of disease-related associations with genetic variants. Watch how the number of associations has increased over time in this video!
Today, the GWAS Catalog contains over 3,400 publications and over 62,000 unique SNP-trait associations. More than 10,000 visitors from around the globe access the GWAS Catalog per month. People using the resource have a wide range of goals – they include researchers wanting to narrow down or prioritise candidate loci, scientists investigating disease mechanisms, clinicians aiming to predict disease risk, pharma industry professionals improving the drug discovery process and anyone wanting to get the latest statistics on disease knowledge or summary data from particular populations of individuals.
The GWAS landscape has evolved over these ten years, with new developments in study design and genotyping technologies. More and more publications on GWAS come out each month. Often a publication contains more than one GWAS – sometimes there are over a thousand in a single paper! The number of individuals studied in a GWAS is also increasing, and so is the number of variants assayed, with bigger arrays and better imputation methods.
The nature of the traits studied is evolving too. Researchers are still publishing GWAS on major common diseases such as type 2 diabetes and breast cancer. However, these studies are now being published alongside studies on ever more specific traits. For example, searching the GWAS Catalog for “schizophrenia” brings up more than 50 different traits related to this disorder. These include sub-phenotypes such as specific symptoms in schizophrenia, endophenotypes such as brain imaging measurements, multi-trait analyses such as “schizophrenia and bipolar disorder”, drug response measurements as well as analyses of the interaction between genetic associations and environmental factors.
All of this means more data to curate. We are adapting our infrastructure constantly to improve curation efficiency so that we can keep up with the rate of data generation. At the same time, we need to maintain the accuracy of the resource and make sure it contains the most relevant and up-to-date research results.
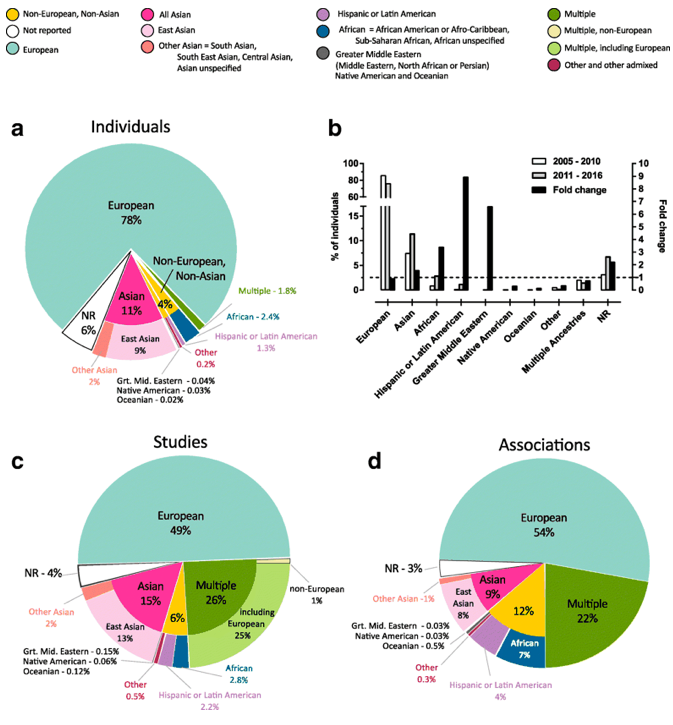
Highlights over the last ten years include releasing a new GWAS Catalog website in 2015, mapping curated trait descriptions to ontology terms to enable enriched ontology-driven search capabilities, improving the interactive GWAS Catalog diagram and capturing ancestry more completely. Accurate characterisation of ancestry is essential to interpret human genomics data. Recently, we have developed a new framework for describing detailed ancestry information and recommendations for reporting ancestry systematically [(Morales et al 2018 Genome Biology 19:2)].(https://europepmc.org/abstract/MED/29448949)
So what will the future hold for the GWAS Catalog? We have just started hosting summary statistics for studies in the GWAS Catalog with support from the scientific community (the files contain the full set of p values for every SNP on the array used). In the future we plan to integrate these datasets with structured meta-data to make it even easier to access GWAS results. We also aim to expand our scope to include other types of association analyses, including targeted arrays and sequencing-based genotyping, which enable deeper interrogation of diseases of interest and inclusion of less common variants.
With these developments come many challenges, including how best to represent the increasing complexity of study designs, trait architecture and statistical analyses. But with the help of the authors of publications in the GWAS Catalog, the scientific community using our resource and our colleagues at Ensembl, we are excited about what the next ten years of GWAS will hold!