📖 PEGASUS Evidence Matrix Introduction
What is the PEGASUS Evidence Matrix?
A PEGASUS Evidence Matrix is the complete, machine-readable table underlying PEG prioritisation. It includes:
- All genes considered at each selected GWAS locus
- Unfiltered values from the relevant evidence categories (not just top-priority genes)
The matrix is designed for pipelines, re-weighting, benchmarking, and cross-study comparison.
PEGASUS Evidence Matrix Requirements
The following standards define the minimal requirements for a PEG Matrix:
-
ONE trait per PEG matrix
- Each matrix corresponds to a single trait or phenotype. -
ONE source GWAS per PEG matrix
- Anchored to a clearly defined GWAS dataset (ideally identified by GWAS Catalog accession ID). -
ONE PEG matrix
- Provided in a machine-readable format (e.g. tab-delimited (TSV); avoid styled or proprietary formats). -
ALL genes at each locus
- Include evidence for every gene considered, not just the top candidate(s).
PEGASUS Evidence Matrix Suggestions (Best Practices)
In addition to the MUST FOLLOW standards above, the following suggestions are recommended to improve interoperability and interpretability of PEG matrices:
-
Evidence summary per gene (author’s conclusion)
- Provide a cumulative “weight of evidence” score or summary statistic. This must be provided if the Matrix is to be represented as a PEGASUS List. -
Standard identifiers
- Use HGNC gene symbols and include Ensembl Gene IDs for unambiguous cross-referencing. -
Avoid evidence double-counting
- When using automated pipelines that integrate multiple evidence sources (e.g., DEPICT, Open Targets), document which sources are included.
- Do not count the same evidence type in one integration analysis twice.
PEGASUS Evidence Matrix Overview
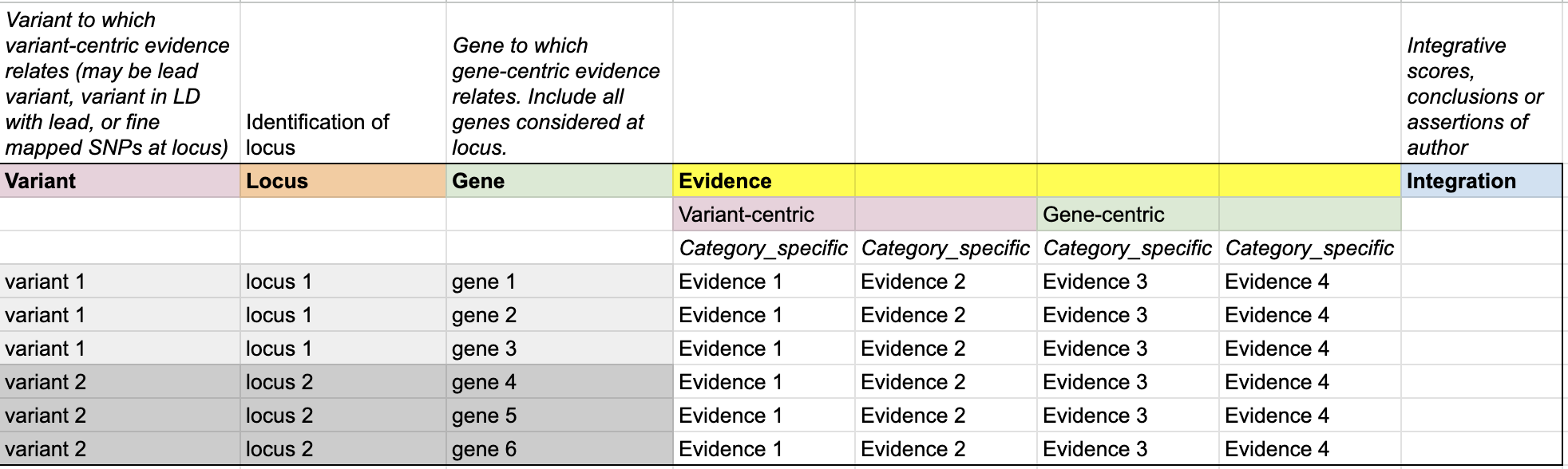
The PEGASUS Evidence Matrix brings together information on genetic variants, locus, and genes, alongside the multiple streams of evidence that connect them. It also includes integrative analyses that based on multiple evidence, as well as a column for conclusions drawn. The structure is designed to make relationships between entities clear and to enable side-by-side comparison of evidence from different sources.
- Variant – Identifies the variant(s) under consideration.
- Locus – Groups variants into locus to provide genomic context.
- Gene – Lists all candidate genes at a locus that are assessed with supporting evidence.
- Evidence – Captures both variant-centric and gene-centric evidence. Evidence is organised into category-specific fields, enabling systematic comparison across genes and loci.
- Integration – Records integrative scores, assertions, or conclusions, combining multiple evidence types into higher-level interpretations.
Together, these components provide a structured and transparent framework for linking genetic signals to potential effector genes, while ensuring the underlying evidence remains visible, comparable, and reproducible.
PEGASUS Evidence Matrix Standard Content
A PEG Evidence Matrix is composed of three major column sections. Together, these define the identifiers, raw evidence, and integrated results for each gene–locus–variant relationship.
Genomic Identifiers
Columns that define which variant, locus, and gene are being studied in each row. These provide the unique indexing needed to anchor all evidence values.
Evidence
Columns that capture the values from individual evidence categories. Each evidence type should be represented in a structured, machine-readable way.
- PEG Evidence Matrix Standard
- Evidence Columns Example
- PEG Metadata Standard
- Metadata Example – Evidence
Integration
Columns that provide summaries or combined scores across multiple evidence categories. These fields make explicit how different sources were weighted, merged, or ranked to prioritise candidate genes.
- PEG Evidence Matrix – Integration
- Integration Columns Example
- PEG Metadata Standard
- Metadata Example – Integration