PEGASUS Framework Overview
Predicted Effector Gene Aggregation, Standards, and Unified Schema (PEGASUS) Framework
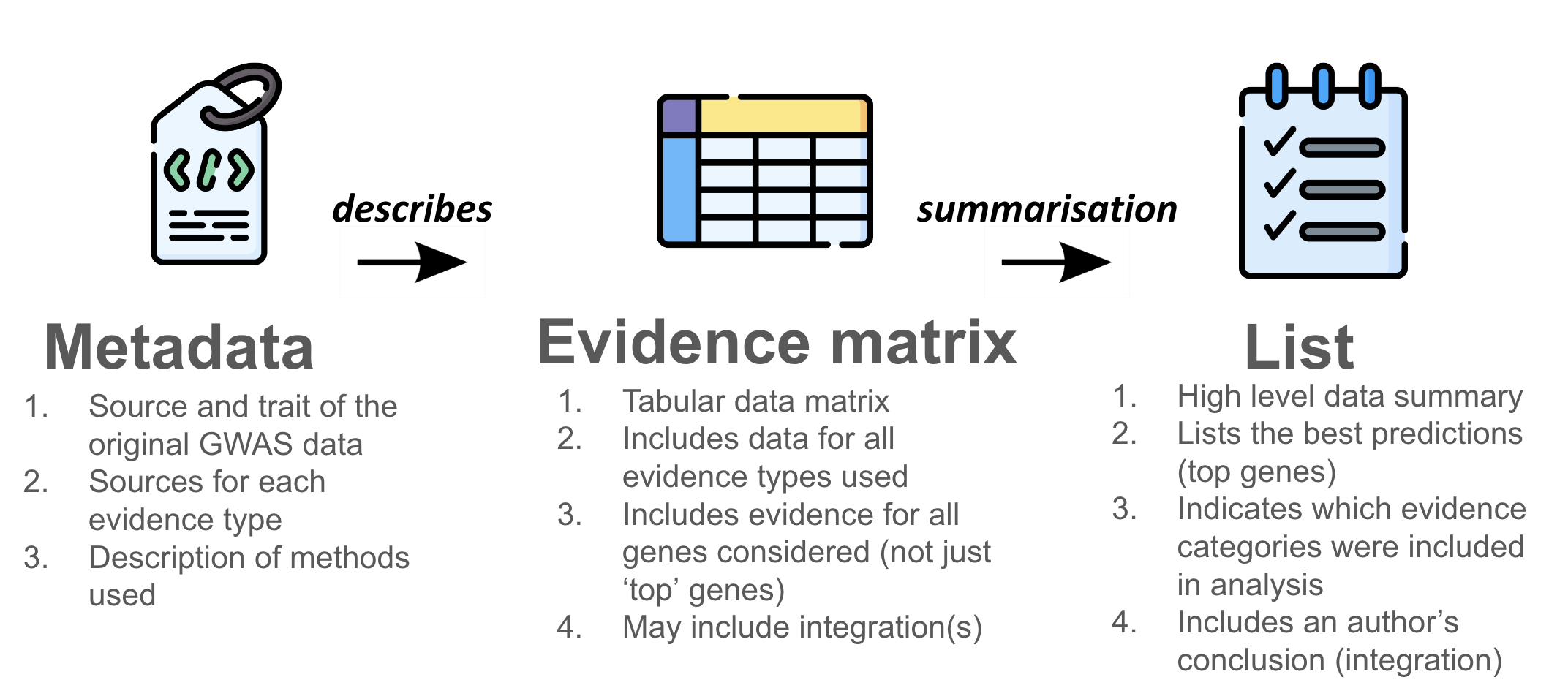
The PEGASUS Framework is a community standard for reporting predicted effector genes derived from genome-wide association studies (GWAS) and integrative evidence. It ensures that results are transparent, interoperable, and reusable by defining three core modules: PEG Metadata, PEG Evidence Matrix and PEG List
-
PEG Evidence Matrix
The full tabular dataset that links variants, loci, and genes as genomic identifiers with all supporting evidence. It includes evidence for all genes (not just top candidates) and may also contain integrated scores.
-
PEG Metadata
Provides the context needed to understand and reuse a PEG matrix, ensuring data are FAIR. It defines column meanings, records data provenance (e.g. biosamples), and specifies methods and integration approaches.
-
PEG List
A summarised output. It highlights the best predictions (top genes), shows which evidence was considered, and includes the author’s conclusion.
PEG Evidence Matrix V.S. PEG Metadata
-
The Evidence Matrix is the data itself — a structured table of variants, genes, and all the evidence linking them. It is what you see and analyse.
-
The Metadata is the description of that data — it explains what each column means, where the data originates, how the data was generated, and how it should be interpreted.
One without the other is incomplete; without the matrix, there is no information to work with; without the metadata, the matrix is hard to interpret and risks being misunderstood. Together, they ensure that PEG data is both rich in content and clear in meaning.
An analogy: The PEG Evidence Matrix and PEG Metadata tell the same story, about the same actors, but from different perspectives. The matrix describes what each actor does in the story, while the metadata reveals their appearance and personality, adding depth and context to their role.
PEG List V.S. PEG Evidence Matrix
-
The Evidence Matrix includes all the evidence for all the genes considered, and includes data for all evidence types used. It may or may not include integrations of different evidence streams.
-
The PEG List is derived from the Evidence Matrix and represents a high level summarisation of the data, showing the top genes only. A PEG List must contain an author’s conclusion, which is an integration of evidence from the Evidence Matrix.
PEG lists:
- Are a concise summary distilled from the PEG Matrix.
- Highlight the top genes most likely causal for the phenotype.
- Summarise which evidence categories were considered, using broad and intuitive categories rather than detailed experimental designs.
PEG Lists must be supported by an Evidence Matrix and described by Metadata.
An analogy:
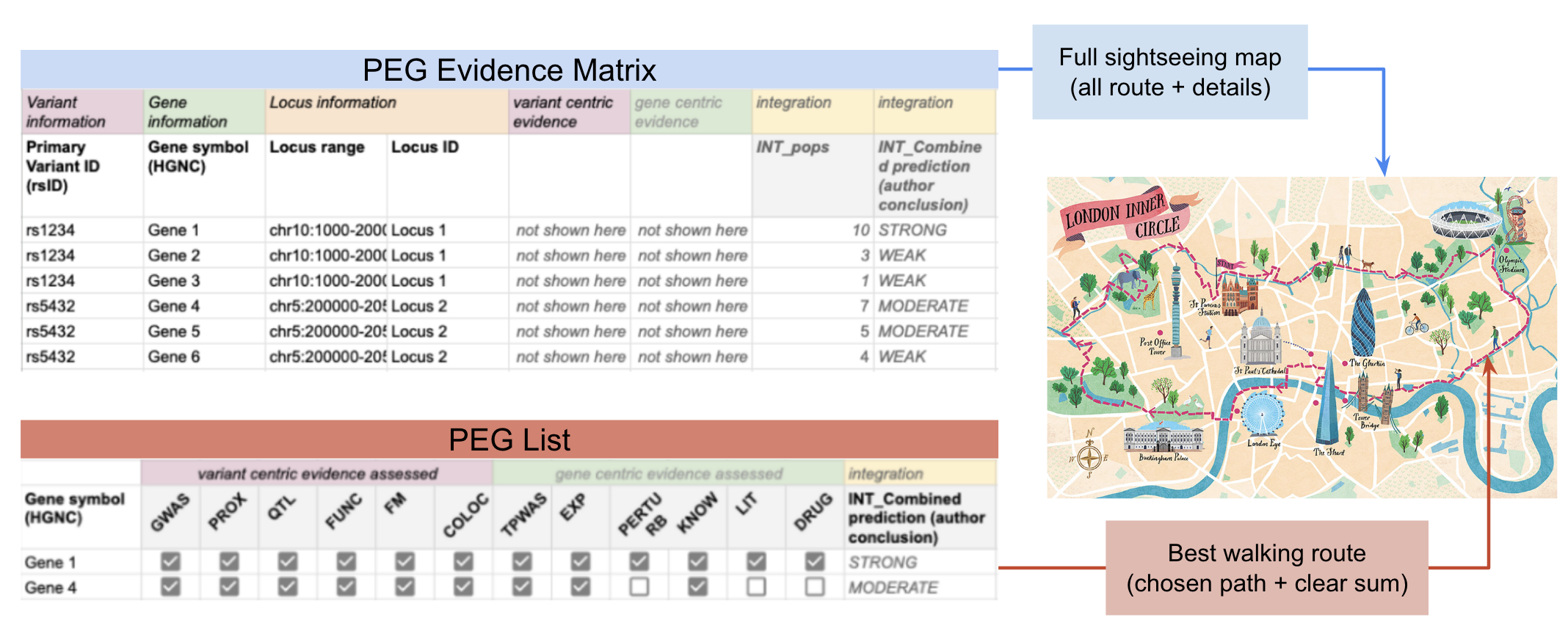
The PEG Evidence Matrix is like a detailed sightseeing map of London.
- It shows all routes, side streets, walking paths, and even underground lines.
- It marks landmarks, hotels, rivers, and attractions — everything you might need to explore the area. This corresponds to the full set of variants, locus, genes, and all evidence categories, including integrated analyses.
The PEG List is like choosing the best walking route for sightseeing.
- From all the possible paths on the map, you highlight the most reasonable route that satisfies your goals (e.g. the author’s criteria). It focuses on the most likely causal genes, supported by the clearest evidence, and provides a concise summary.