Toy Data (PEGASUS Framework applied)🎠
Toy data
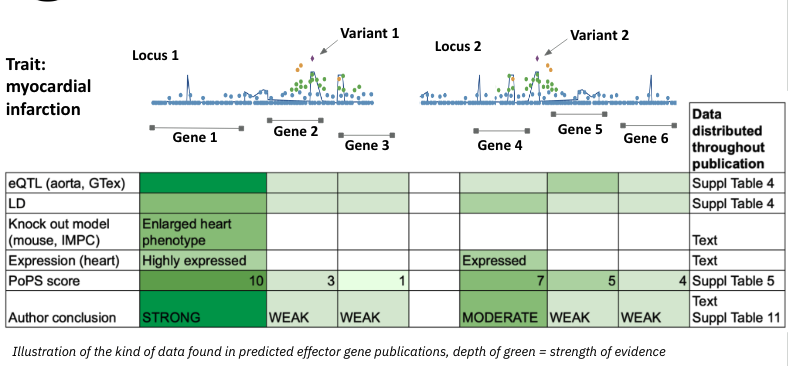
This toy data shows two loci identified by lead associations from a GWAS for the trait myocardial infarction. Each locus contains multiple nearby candidate effector genes (Gene 1–6).
The bottom table summarises the supporting evidence for each gene — including eQTLs, predicted functional impact, gene expression in aorta, gene prioritisation scores generated by PoPS, and the authors’ overall conclusion.
Importantly, in publications this type of evidence is often scattered across the main text and multiple supplementary tables, making it difficult to compare, integrate, or reproduce.
PEGASUS Evidence Matrix
In the PEGASUS Evidence Matrix, we propose presenting all evidence in a single structured table. The following table illustrates how the same information can be reformatted into a unified matrix.
| Primary Variant ID | rsID | Gene ID | Gene symbol | Locus range | Locus ID | GWAS_pvalue | FUNC_CADD | QTL_eQTL_aorta_pvalue | EXP_aorta_RPKM | PERTURB_mouse | INT_pops | INT_Combined prediction (author score) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr1:100000:T:C | rs1234 | ENSG00000000001 | Gene 1 | chr1:99500-115000 | rs1234 | 4.00E-09 | 18.2 | 7.00E-07 | 8.7 | enlarged heart | increased heart weight | 10 | STRONG |
| chr1:100000:T:C | rs1234 | ENSG00000000002 | Gene 2 | chr1:99500-115000 | rs1234 | 4.00E-09 | 3.45 | 0.01 | NA | NA | 3 | WEAK |
| chr1:100000:T:C | rs1234 | ENSG00000000003 | Gene 3 | chr1:99500-115000 | rs1234 | 4.00E-09 | 6.4 | 0.05 | NA | NA | 1 | WEAK |
| chr2:20000:A:G | rs5432 | ENSG00000000004 | Gene 4 | chr2:19000-21000 | rs5432 | 3.00E-08 | 15.62 | 8.00E-05 | 1.3 | NA | 7 | MODERATE |
| chr2:20000:A:G | rs5432 | ENSG00000000005 | Gene 5 | chr2:19000-21000 | rs5432 | 3.00E-08 | 2.13 | 0.2 | NA | NA | 5 | WEAK |
| chr2:20000:A:G | rs5432 | ENSG00000000006 | Gene 6 | chr2:19000-21000 | rs5432 | 3.00E-08 | 4.4 | 0.05 | NA | NA | 4 | WEAK |
PEGASUS List
The PEGASUS List distils the matrix into a concise summary, highlighting the strongest candidate gene at each locus. The PEGASUS List Foundational model records whether evidence was considered (tick = data present, blank = not assessed) and reflects the author’s integrated conclusions for top genes.
| rsID | Gene symbol | Variant-centric | Gene-centric | INT_Combined prediction (author score) | |||
|---|---|---|---|---|---|---|---|
| GWAS | FUNC | QTL | EXP | PERTUB | |||
| rs1234 | Gene 1 | STRONG | |||||
| rs5432 | Gene 4 | MODERATE | |||||
Tick = data/value present (VAL). Blank = not assessed (NA). Ticks do not imply supportive vs negative; see author interpretation & provenance
PEGASUS Metadata
PEGASUS Metadata — Provides the detailed context behind the PEGASUS Matrix, recording column definitions, provenance, biosamples, and methods so that PEG evidence is fully interpretable and reproducible.
PEGASUS Metadata in Excel (suitable for submission)
- 📂 Dataset description
- 🧬 Genomic Identifier tab
- 🔎 Evidence tab
- 🔗 Integration tab
| peg_source | gwas_source | trait_description | trait_ontology_id | sample_description | sample_size | case_control_study | sample_ancestry | sample_ancestry_label |
|---|---|---|---|---|---|---|---|---|
| PMID:36357675 | PMID:36357675 | Ascorbic acid 3-sulfate levels | EFO_0800173 | 6,136 Finnish ancestry individuals | 6136 | False | Finland | European |
| variant_type | genome_build | variant_information | gene_id_source_version | gene_symbol_source_version | info | locus_type | locus_id | locus_info |
|---|---|---|---|---|---|---|---|---|
| lead | GRCh38 | The primary variant is the variant with the most significant association p-value in the study | Ensembl v109 | HGNC 2025-07-30 | NA | LD | Lead SNP | NA |
| column_header | column_description | stream_name | category | category_abbreviation | class | source_tag | method_tag | threshold | notes |
|---|---|---|---|---|---|---|---|---|---|
| GWAS_pvalue | Association p-value for each variant from the GWAS study | GWAS | Genome-wide association (GWAS) signal | GWAS | variant-centric | NA | NA | NA | NA |
| FUNC_CADD | CADD score representing the predicted functional impact of the variant | FUNC | Predicted functional impact | FUNC | variant-centric | source_cadd | NA | NA | NA |
| QTL_eQTL_aorta_pvalue | p-value from expression QTL (eQTL) analysis in aorta tissue | eQTL | Molecular QTL | QTL | variant-centric | source_gtex_aorta_qtl | soft_fastqtl | qvalue < 0.05 | NA |
| EXP_aorta_RPKM | Gene expression level in aorta tissue, measured in RPKM | EXP | Expression | EXP | gene-centric | source_gtex_aorta_rna | NA | NA | NA |
| PERTURB_mouse | Phenotypic effects of the gene from IMPC knockout mouse models | PERTURB | Perturbation | PERTURB | gene-centric | source_impc | NA | NA | NA |
| column_header | column_description | integration_analysis | evidence_stream_name | integrated_analysis_name | method_tag | threshold | notes |
|---|---|---|---|---|---|---|---|
| INT_pops | Integrated score based on multiple evidence types for the prioritised gene | pops | FUNC | eQTL | pQTL | FM | 3D | PHEWAS | TWAS | NA | soft_pops | score > 3 | NA |
| INT_Combined prediction (author score) | Combined prediction based on manual review of all evidence types and PoPS output | author_score | PROX | REG | LIT | PoPS | pops | method_customised | NA | NA |
- 📚 Source tab
- ⚙️ Method tab
| source_tag | provenance | file_name | version | url | accesstion | doi | tissue | sample_origin | cell_type | cell_line | disease | life_stage | treatment | sex | age | species | description |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| source_cadd | CADD | All possible SNVs of GRCh38/hg38 incl. all annotations | v1.7 | link | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| source_gtex_eqtl | GTEx | GTEx_Analysis_v10_eQTL.tar | v10 | link | NA | NA | aorta | primary tissue | NA | NA | healthy | adult | None | mixed | mixed | Homo sapiens | Bulk aorta tissueSamples from healthy adult human donors in GTEx v10. Used for eQTL discovery. Donors aged ~20–70 years, male and female. |
| source_gtex_aorta_RNA | GTEx | GTEx_Analysis_v10_RNASeQCv2.4.2_gene_tpm.gct.gz | v10 | link | NA | NA | aorta | primary tissue | NA | NA | healthy | adult | None | mixed | mixed | Homo sapiens | Bulk aorta tissuesamples (GTEx v10) from healthy postmortem adult human donors in GTEx v10. Used for RNA expression profiling. Donors aged ~20–70 years, male and female. |
| source_impc | IMPC | IMPC_genotype_phenotype.csv.gz | 23 | link | NA | NA | multiple | IMPC mouse knockout models | NA | NA | NA | mixed | gene knockout | mixed | mixed | Mus musculus | Mice with single-gene knockouts generated by the IMPC project. |
| method_tag | method_mode | software_name | software_version | software_url | software_doi | method_description |
|---|---|---|---|---|---|---|
| soft_fastqtl | computational | FastQTL | v1.0 | link | 10.1093/BIOINFORMATICS/BTV722 | NA |
| soft_pops | computational | PoPS | v1.0 | link | 10.1038/s41588-023-01443-6 | NA |
| method_customise | manual | NA | NA | NA | NA | An integrated predictionderived from expert review of all evidence types together with PoPS output. The strength of support for a gene is classified as weak, medium, or strong based on professional judgement: |
PEGASUS Metadata in YAML (suitable for reader)
Using YAML for metadata keeps all information on one page in a structured format, so users can easily search and extract the details they need