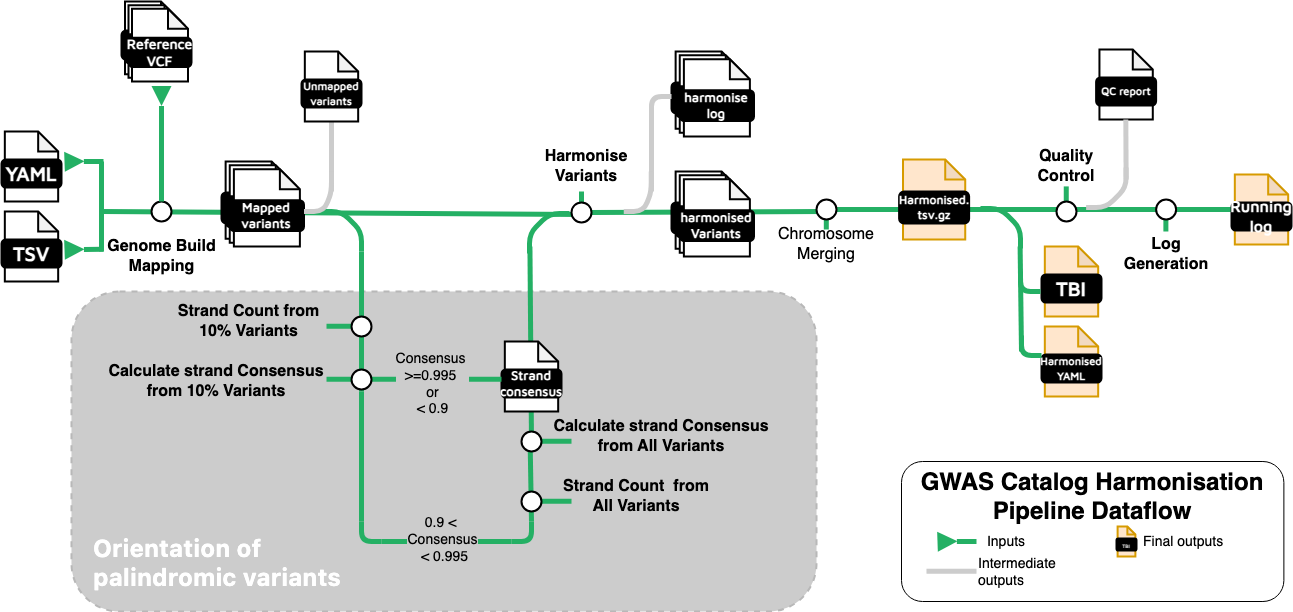
Overview of the Harmonisation Pipeline
The gwas-sumstats-harmoniser is a pipeline designed to standardise variant data across different genome assemblies, ensuring consistency for downstream analysis. This process involves four key steps:
-
Genome Build Mapping: Lifting variants to the desired genome assembly (GRCh38) to align genomic positions to a unified coordinate system.
-
Palindromic Variant Orientation: Inferring strand orientation of palindromic variants using a strand consensus approach.
-
Variant Harmonisation: Matching and aligning variants with those in a reference dataset to ensure allele consistency and orientation to the forward strand.
-
Quality control: Removing variants missing any essential column value (chromosome, base pair location, or p-value).